MacroAPE 1083:CGGAAACGGAA,motif609
From FANTOM5_SSTAR
Full Name: CGGAAACGGAA,motif609,CNhs12591,T:337.0(15.06%),B:5266.2(7.76%),P:1e-30
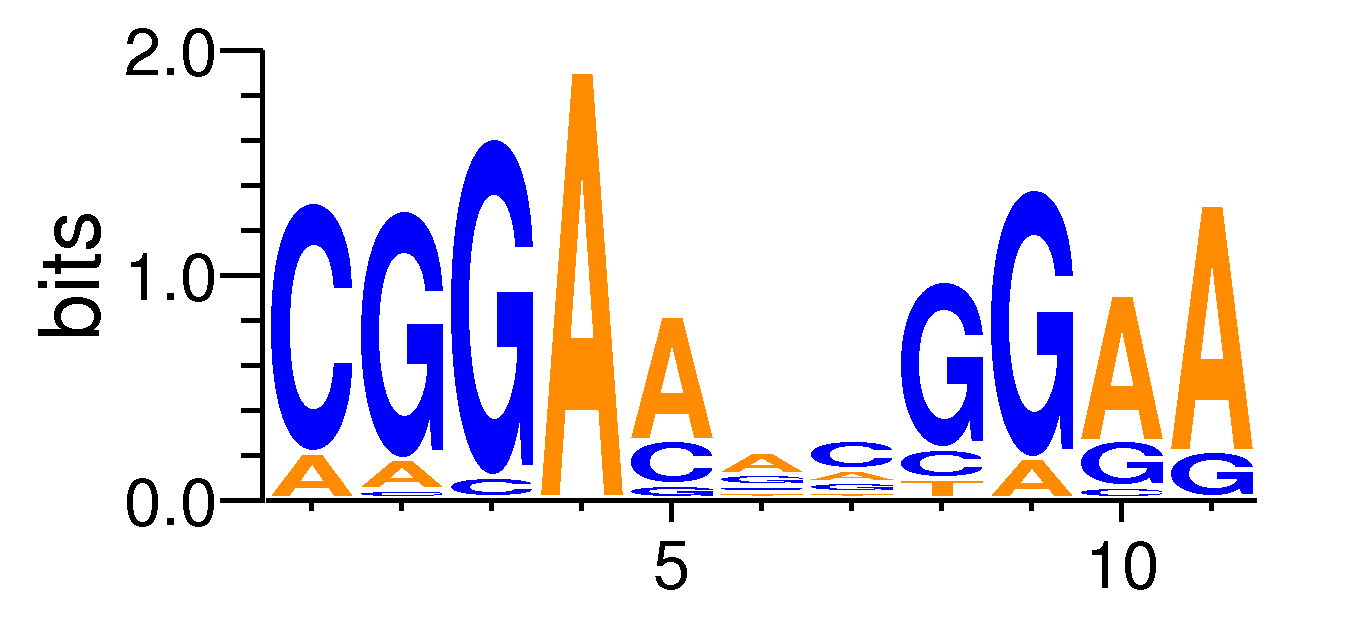
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| id1#MA0120.1 | 0 | 0.00017325 | 0.0824668 | 0.109164 | 11 | CGGAAACGGAA | CGAAAAGGAAAA | - |
| HAL9#MA0311.1 | 0 | 0.000231348 | 0.110122 | 0.109164 | 5 | CGGAAACGGAA | CGGAA | + |
| ETS1#MA0098.1 | 0 | 0.000354716 | 0.168845 | 0.111584 | 6 | CGGAAACGGAA | CGGAAA | - |
| GABPA#MA0062.2 | 1 | 0.000694918 | 0.330781 | 0.133495 | 10 | CGGAAACGGAA | CCGGAAGTGGC | + |
| STAT1#MA0137.1 | -1 | 0.000820088 | 0.390362 | 0.133495 | 10 | CGGAAACGGAA | GGAAAACGAAACTG | + |
| LEU3#MA0324.1 | 1 | 0.00084874 | 0.404 | 0.133495 | 9 | CGGAAACGGAA | CCGGTAACGG | - |
| Eip74EF#MA0026.1 | 1 | 0.00141182 | 0.672028 | 0.190338 | 6 | CGGAAACGGAA | CCGGAAG | + |
| SPI1#MA0080.1 | 0 | 0.00296348 | 1.41061 | 0.290675 | 6 | CGGAAACGGAA | CGGAAG | + |
| CAT8#MA0280.1 | 1 | 0.00296348 | 1.41061 | 0.290675 | 5 | CGGAAACGGAA | CCGGAA | + |
| CEP3#MA0282.1 | 2 | 0.00308011 | 1.46613 | 0.290675 | 6 | CGGAAACGGAA | CTCGGAAA | + |
| ASG1#MA0275.1 | 1 | 0.00363664 | 1.73104 | 0.311997 | 5 | CGGAAACGGAA | CCGGAA | + |
| TBS1#MA0404.1 | 0 | 0.00420751 | 2.00278 | 0.330892 | 8 | CGGAAACGGAA | CGGATCCG | + |
| Su(H)#MA0085.1 | 4 | 0.00467605 | 2.2258 | 0.339452 | 11 | CGGAAACGGAA | CTGTGGGAAACGAGAT | + |
| STB4#MA0391.1 | 2 | 0.0052098 | 2.47986 | 0.351185 | 5 | CGGAAACGGAA | CTCGGAA | + |
| ELK1#MA0028.1 | -2 | 0.00589244 | 2.8048 | 0.355164 | 9 | CGGAAACGGAA | GAGCCGGAAG | + |
| IXR1#MA0323.1 | 4 | 0.00622602 | 2.96359 | 0.355164 | 11 | CGGAAACGGAA | AAGCCGGAAGCGGGG | + |
| GABPA#MA0062.2 | 2 | 0.00639787 | 3.04539 | 0.355164 | 8 | CGGAAACGGAA | ACCGGAAGAG | + |
| SPIB#MA0081.1 | -4 | 0.00776235 | 3.69488 | 0.39758 | 7 | CGGAAACGGAA | AGAGGAA | + |
| YRM1#MA0438.1 | 1 | 0.00803417 | 3.82427 | 0.39758 | 7 | CGGAAACGGAA | ACGGAAAT | + |
| RDR1#MA0360.1 | 2 | 0.00842581 | 4.01069 | 0.39758 | 6 | CGGAAACGGAA | TGCGGAAC | + |
| che-1#MA0260.1 | -2 | 0.0113762 | 5.41505 | 0.494951 | 6 | CGGAAACGGAA | GAAACC | + |
| IRF2#MA0051.1 | -1 | 0.0115383 | 5.49224 | 0.494951 | 10 | CGGAAACGGAA | GGAAAGCGAAACCAAAAC | + |
| YKL222C#MA0428.1 | -4 | 0.016078 | 7.65312 | 0.623001 | 7 | CGGAAACGGAA | AACGGAAAT | + |
| Pax5#MA0014.1 | 6 | 0.0165039 | 7.85584 | 0.623001 | 11 | CGGAAACGGAA | GGAGCACTGAAGCGTAACCG | + |
| SIP4#MA0380.1 | 2 | 0.0189958 | 9.042 | 0.663266 | 5 | CGGAAACGGAA | TCCGGAG | - |
| IRF1#MA0050.1 | -2 | 0.0206865 | 9.84678 | 0.663266 | 9 | CGGAAACGGAA | GAAAGCGAAACC | + |