MacroAPE 1083:Motif37
From FANTOM5_SSTAR
Full Name: motif37
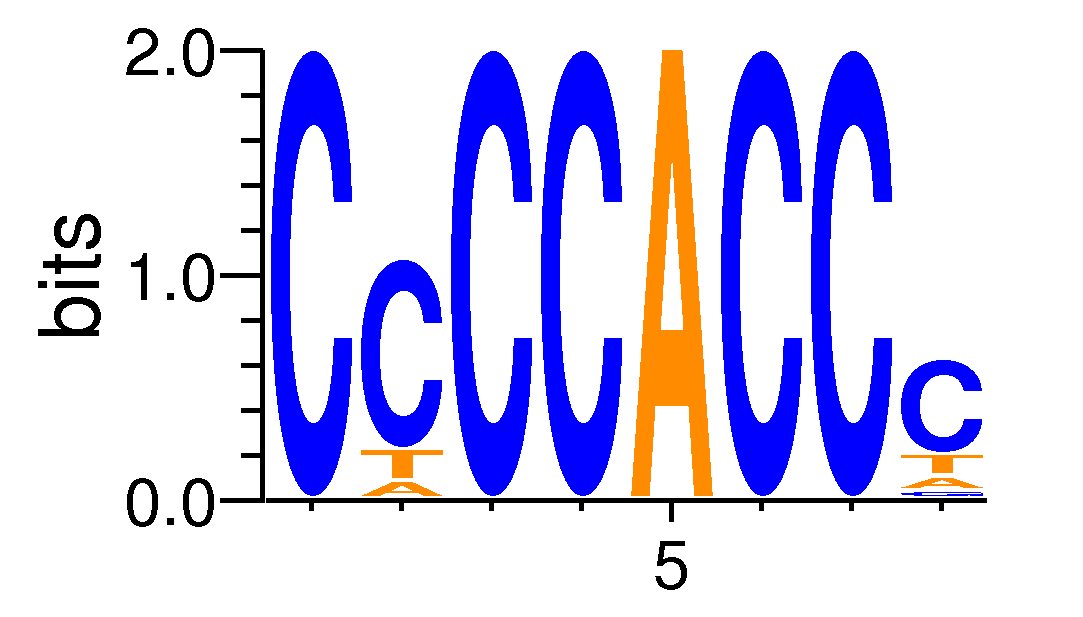
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Klf4#MA0039.2 | 1 | 2.67455e-05 | 0.0127308 | 0.0252402 | 8 | CCCCACCC | GCCCCACCCA | - |
| YPR022C#MA0436.1 | 0 | 6.83407e-05 | 0.0325302 | 0.0322472 | 7 | CCCCACCC | CCCCACG | + |
| SP1#MA0079.2 | 0 | 0.000164423 | 0.0782654 | 0.051723 | 8 | CCCCACCC | CCCCGCCCCC | + |
| SP1#MA0079.2 | 1 | 0.000460496 | 0.219196 | 0.0755257 | 8 | CCCCACCC | ACCCCGCCCC | - |
| MIG2#MA0338.1 | 0 | 0.00048977 | 0.233131 | 0.0755257 | 7 | CCCCACCC | CCCCGCA | + |
| MIG3#MA0339.1 | 0 | 0.000551026 | 0.262289 | 0.0755257 | 7 | CCCCACCC | CCCCGCA | + |
| RPN4#MA0373.1 | 0 | 0.000648437 | 0.308656 | 0.0755257 | 7 | CCCCACCC | CGCCACC | - |
| YGR067C#MA0425.1 | 1 | 0.000655667 | 0.312098 | 0.0755257 | 8 | CCCCACCC | ACCCCACTTTTTTG | + |
| YML081W#MA0431.1 | 1 | 0.000720269 | 0.342848 | 0.0755257 | 8 | CCCCACCC | ACCCCGCAC | + |
| ADR1#MA0268.1 | 1 | 0.000833525 | 0.396758 | 0.0786613 | 6 | CCCCACCC | ACCCCAC | + |
| RREB1#MA0073.1 | 11 | 0.00153708 | 0.731652 | 0.13187 | 8 | CCCCACCC | CCCCAAACCACCCCCCCCCC | + |
| MIG1#MA0337.1 | 1 | 0.00215186 | 1.02429 | 0.169229 | 6 | CCCCACCC | CCCCCGC | + |
| ZNF354C#MA0130.1 | 0 | 0.0046922 | 2.23349 | 0.340625 | 6 | CCCCACCC | ATCCAC | + |
| AFT2#MA0270.1 | -1 | 0.00526051 | 2.504 | 0.354603 | 7 | CCCCACCC | CACACCCC | + |
| MET31#MA0333.1 | 0 | 0.00594033 | 2.8276 | 0.373733 | 8 | CCCCACCC | CGCCACACT | - |
| MZF1_1-4#MA0056.1 | 1 | 0.0087643 | 4.17181 | 0.516939 | 5 | CCCCACCC | TCCCCA | - |
| CHA4#MA0283.1 | 1 | 0.00990658 | 4.71553 | 0.549943 | 7 | CCCCACCC | TCTCCGCC | - |
| ASH1#MA0276.1 | 0 | 0.0112691 | 5.36409 | 0.580822 | 8 | CCCCACCC | CCCGATCCGG | - |
| Macho-1#MA0118.1 | 0 | 0.0116938 | 5.56623 | 0.580822 | 8 | CCCCACCC | GAACCCCCA | - |
| MET32#MA0334.1 | 0 | 0.0130445 | 6.2092 | 0.605457 | 7 | CCCCACCC | CGCCACA | + |
| ZMS1#MA0441.1 | 2 | 0.0138163 | 6.57654 | 0.605457 | 7 | CCCCACCC | TTCCCCGCA | + |
| SUT1#MA0399.1 | 0 | 0.0141144 | 6.71848 | 0.605457 | 7 | CCCCACCC | CCCCGCG | - |
| Egr1#MA0162.1 | 2 | 0.0152815 | 7.27399 | 0.606779 | 8 | CCCCACCC | CCGCCCACGCA | - |
| MZF1_5-13#MA0057.1 | 2 | 0.0158423 | 7.54095 | 0.606779 | 8 | CCCCACCC | TTCCCCCTAC | - |
| opa#MA0456.1 | 2 | 0.0160742 | 7.6513 | 0.606779 | 8 | CCCCACCC | GACCCCCCGCTG | + |
| Pax4#MA0068.1 | 21 | 0.0168285 | 8.01038 | 0.610823 | 8 | CCCCACCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| btd#MA0443.1 | 1 | 0.0192735 | 9.17421 | 0.661995 | 8 | CCCCACCC | TCCGCCCCCT | - |
| UGA3#MA0410.1 | 0 | 0.0196413 | 9.34926 | 0.661995 | 8 | CCCCACCC | TCCCGCCG | - |