MacroAPE 1083:STAT4 CD4-Stat4-ChIP-Seq/Homer
From FANTOM5_SSTAR
Full Name: STAT4_CD4-Stat4-ChIP-Seq/Homer
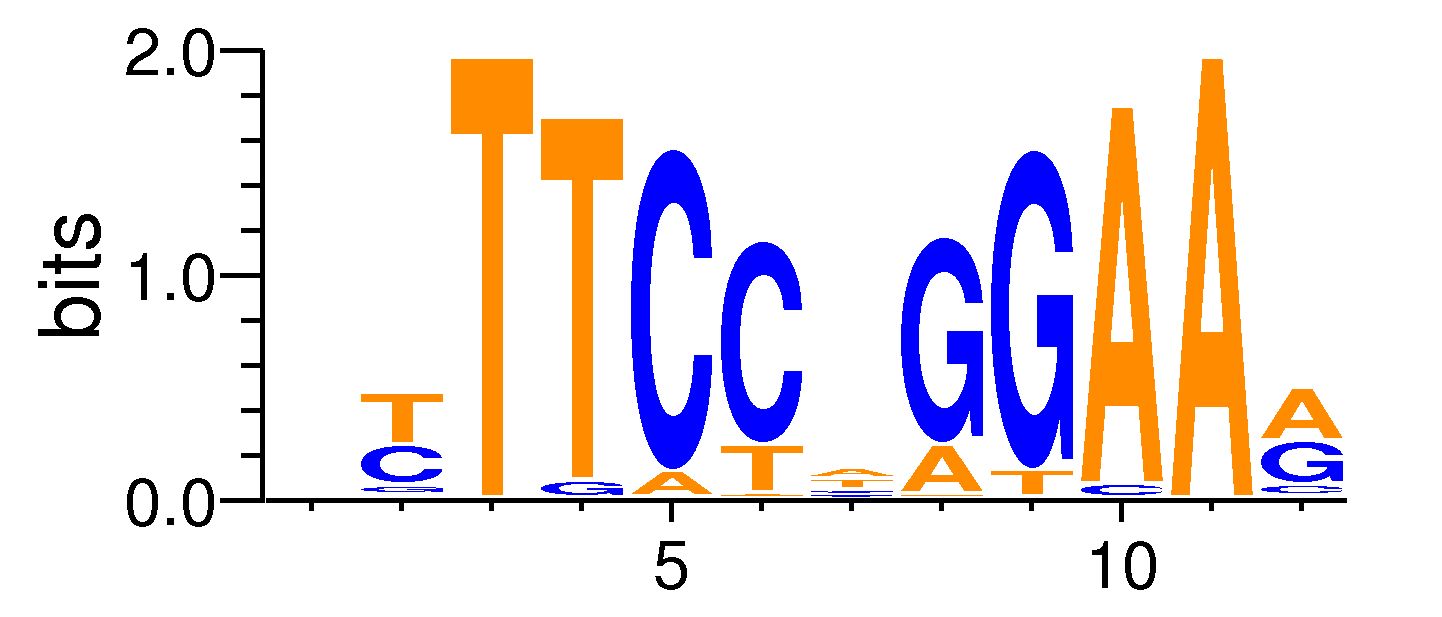
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| STAT1#MA0137.2 | 1 | 4.97216e-11 | 2.36675e-08 | 4.7273e-08 | 12 | GTTTCCAGGAAA | GGTTTCCGGGAAATG | - |
| Stat3#MA0144.1 | -2 | 2.37372e-08 | 1.12989e-05 | 7.17666e-06 | 10 | GTTTCCAGGAAA | TTCCAGGAAG | + |
| Eip74EF#MA0026.1 | -1 | 0.00366944 | 1.74665 | 0.697746 | 7 | GTTTCCAGGAAA | CTTCCGG | - |
| FEV#MA0156.1 | -5 | 0.00523855 | 2.49355 | 0.71151 | 7 | GTTTCCAGGAAA | CAGGAAAT | + |
| ETS1#MA0098.1 | -1 | 0.0071763 | 3.41592 | 0.7581 | 6 | GTTTCCAGGAAA | CTTCCG | + |
| ELK1#MA0028.1 | -1 | 0.013483 | 6.41789 | 0.998691 | 10 | GTTTCCAGGAAA | CTTCCGGCTC | - |
| PUT3#MA0358.1 | -3 | 0.0139285 | 6.62999 | 0.998691 | 8 | GTTTCCAGGAAA | CCCGGGAA | + |
| GCR2#MA0305.1 | 0 | 0.0161 | 7.66358 | 0.998691 | 7 | GTTTCCAGGAAA | GCTTCCT | + |
| SPI1#MA0080.1 | -6 | 0.0199359 | 9.48949 | 0.998691 | 6 | GTTTCCAGGAAA | CGGAAG | + |
| ASG1#MA0275.1 | -2 | 0.0199359 | 9.48949 | 0.998691 | 6 | GTTTCCAGGAAA | TTCCGG | - |