MacroAPE 1083:NANOG mES-Nanog-ChIP-Seq/Homer
From FANTOM5_SSTAR
Full Name: NANOG_mES-Nanog-ChIP-Seq/Homer
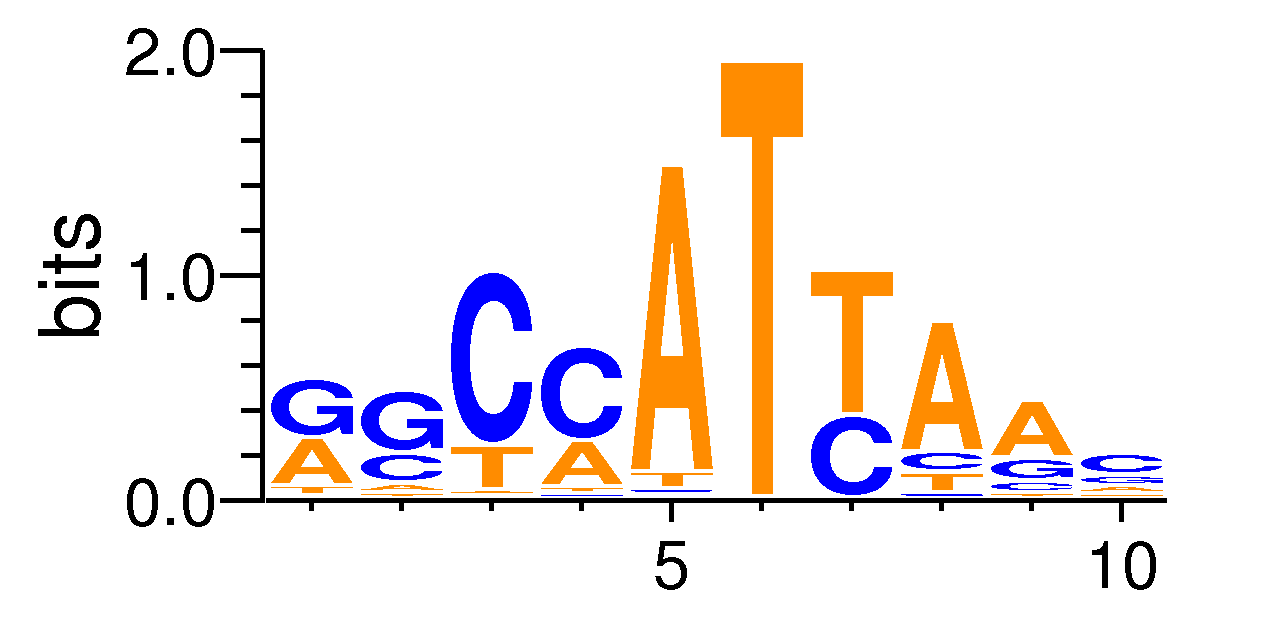
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| tup#MA0248.1 | -2 | 9.48507e-06 | 0.00451489 | 0.00850286 | 7 | GGCCATTAAC | CAATTAA | - |
| bsh#MA0214.1 | -2 | 3.1417e-05 | 0.0149545 | 0.0140818 | 7 | GGCCATTAAC | CAATTAA | - |
| NK7.1#MA0196.1 | -2 | 0.000160699 | 0.0764928 | 0.0319138 | 7 | GGCCATTAAC | CAATTAA | - |
| CG11085#MA0171.1 | -2 | 0.000178002 | 0.0847289 | 0.0319138 | 7 | GGCCATTAAC | CAATTAA | - |
| CG13424#MA0175.1 | -2 | 0.000178002 | 0.0847289 | 0.0319138 | 7 | GGCCATTAAC | CAATTAA | - |
| slou#MA0245.1 | -2 | 0.000239539 | 0.114021 | 0.0337797 | 7 | GGCCATTAAC | CAATTAA | - |
| ems#MA0219.1 | -2 | 0.000263773 | 0.125556 | 0.0337797 | 7 | GGCCATTAAC | TCATTAA | - |
| C15#MA0170.1 | -2 | 0.000461146 | 0.219506 | 0.0421279 | 7 | GGCCATTAAC | TAATTAA | - |
| Nobox#MA0125.1 | 0 | 0.000575405 | 0.273893 | 0.0421279 | 8 | GGCCATTAAC | ACCAATTA | - |
| unc-4#MA0250.1 | -2 | 0.000602697 | 0.286884 | 0.0421279 | 7 | GGCCATTAAC | CAATTAA | - |
| B-H2#MA0169.1 | -2 | 0.000657921 | 0.31317 | 0.0421279 | 7 | GGCCATTAAC | CAATTAA | - |
| CG15696#MA0176.1 | -2 | 0.000657921 | 0.31317 | 0.0421279 | 7 | GGCCATTAAC | CAATTAA | - |
| Hmx#MA0192.1 | -2 | 0.000657921 | 0.31317 | 0.0421279 | 7 | GGCCATTAAC | CAATTAA | - |
| ftz#MA0225.1 | -2 | 0.000657921 | 0.31317 | 0.0421279 | 7 | GGCCATTAAC | TAATTAA | - |
| Dll#MA0187.1 | -1 | 0.00071717 | 0.341373 | 0.0428603 | 7 | GGCCATTAAC | GTAATTA | - |
| CG34031#MA0444.1 | -2 | 0.00078046 | 0.371499 | 0.0437276 | 7 | GGCCATTAAC | CAATTAA | - |
| Dfd#MA0186.1 | -2 | 0.000920479 | 0.438148 | 0.0461553 | 7 | GGCCATTAAC | TCATTAA | - |
| CG32532#MA0179.1 | -2 | 0.000998053 | 0.475073 | 0.0461553 | 7 | GGCCATTAAC | TAATTAA | - |
| btn#MA0215.1 | -2 | 0.000998053 | 0.475073 | 0.0461553 | 7 | GGCCATTAAC | TCATTAA | - |
| E5#MA0189.1 | -2 | 0.00108123 | 0.514664 | 0.0461553 | 7 | GGCCATTAAC | TAATTAA | - |
| eve#MA0221.1 | -2 | 0.00108123 | 0.514664 | 0.0461553 | 7 | GGCCATTAAC | TCATTAG | - |
| Scr#MA0203.1 | -2 | 0.00117002 | 0.556929 | 0.0476755 | 7 | GGCCATTAAC | TCATTAA | - |
| HGTX#MA0191.1 | -2 | 0.00136315 | 0.648861 | 0.0509164 | 7 | GGCCATTAAC | TAATTAA | - |
| en#MA0220.1 | -2 | 0.00136315 | 0.648861 | 0.0509164 | 7 | GGCCATTAAC | TAATTAA | - |
| Ubx#MA0094.2 | -2 | 0.00144744 | 0.688979 | 0.0519019 | 8 | GGCCATTAAC | TAATTAAA | - |
| B-H1#MA0168.1 | -2 | 0.00158044 | 0.75229 | 0.0524734 | 7 | GGCCATTAAC | CAATTAA | - |
| zen2#MA0257.1 | -2 | 0.00158044 | 0.75229 | 0.0524734 | 7 | GGCCATTAAC | TAATTAA | - |
| Antp#MA0166.1 | -2 | 0.00169919 | 0.808814 | 0.0525252 | 7 | GGCCATTAAC | TCATTAA | - |
| Dr#MA0188.1 | -1 | 0.00169919 | 0.808814 | 0.0525252 | 7 | GGCCATTAAC | CCAATTA | + |
| hbn#MA0226.1 | -2 | 0.00195917 | 0.932563 | 0.0585429 | 7 | GGCCATTAAC | TAATTAA | - |
| unpg#MA0251.1 | -2 | 0.00225306 | 1.07246 | 0.0651531 | 7 | GGCCATTAAC | TAATTAA | - |
| Lim3#MA0195.1 | -2 | 0.00241472 | 1.1494 | 0.0676458 | 7 | GGCCATTAAC | TAATTAA | - |
| Vsx1#MA0181.1 | -2 | 0.00258731 | 1.23156 | 0.0695383 | 7 | GGCCATTAAC | TAATTAA | - |
| Oct#MA0197.1 | -2 | 0.00263741 | 1.25541 | 0.0695383 | 8 | GGCCATTAAC | TAATTAAA | - |
| abd-A#MA0206.1 | -2 | 0.00296931 | 1.41339 | 0.0760522 | 7 | GGCCATTAAC | TAATTAA | - |
| Vsx2#MA0180.1 | -1 | 0.00335439 | 1.59669 | 0.0822069 | 9 | GGCCATTAAC | CTAATTAAA | - |
| otp#MA0236.1 | -2 | 0.00364965 | 1.73723 | 0.0822069 | 7 | GGCCATTAAC | TAATTAA | - |
| pb#MA0238.1 | -2 | 0.00364965 | 1.73723 | 0.0822069 | 7 | GGCCATTAAC | TAATTAA | - |
| YY1#MA0095.1 | -1 | 0.00366812 | 1.74603 | 0.0822069 | 6 | GGCCATTAAC | GCCATC | + |
| YHP1#MA0426.1 | -2 | 0.00366812 | 1.74603 | 0.0822069 | 6 | GGCCATTAAC | CAATTA | - |
| lab#MA0230.1 | -2 | 0.00390917 | 1.86077 | 0.0834372 | 7 | GGCCATTAAC | TAATTAA | - |
| H2.0#MA0448.1 | -2 | 0.00390917 | 1.86077 | 0.0834372 | 7 | GGCCATTAAC | TAATTAA | - |
| Awh#MA0167.1 | -2 | 0.00448454 | 2.13464 | 0.0886059 | 7 | GGCCATTAAC | TAATTAA | - |
| CG18599#MA0177.1 | -2 | 0.00448454 | 2.13464 | 0.0886059 | 7 | GGCCATTAAC | TAATTAA | - |
| Rx#MA0202.1 | -2 | 0.00480176 | 2.28564 | 0.0886059 | 7 | GGCCATTAAC | TAATTAG | - |
| ap#MA0209.1 | -2 | 0.00480176 | 2.28564 | 0.0886059 | 7 | GGCCATTAAC | TAATTAG | - |
| ind#MA0228.1 | -2 | 0.00480176 | 2.28564 | 0.0886059 | 7 | GGCCATTAAC | TAATTAG | - |
| lbl#MA0232.1 | -2 | 0.00497306 | 2.36718 | 0.0886059 | 6 | GGCCATTAAC | TAATTA | - |
| Ubx#MA0094.2 | -4 | 0.00504727 | 2.4025 | 0.0886059 | 4 | GGCCATTAAC | ATTA | - |
| CG42234#MA0174.1 | -2 | 0.00513974 | 2.44652 | 0.0886059 | 7 | GGCCATTAAC | TAATAAA | - |
| zen#MA0256.1 | -2 | 0.00513974 | 2.44652 | 0.0886059 | 7 | GGCCATTAAC | TCATTAG | - |
| PHDP#MA0457.1 | -2 | 0.00513974 | 2.44652 | 0.0886059 | 7 | GGCCATTAAC | TAATTAA | - |
| Nkx2-5#MA0063.1 | -2 | 0.00549985 | 2.61793 | 0.0930249 | 7 | GGCCATTAAC | CAATTAA | - |
| HAP4#MA0315.1 | 0 | 0.00575231 | 2.7381 | 0.094185 | 10 | GGCCATTAAC | ACCAATCAGAGGCGA | + |
| OdsH#MA0455.1 | -2 | 0.00588363 | 2.80061 | 0.094185 | 7 | GGCCATTAAC | TAATTAG | - |
| OdsH#MA0455.1 | -2 | 0.00588363 | 2.80061 | 0.094185 | 7 | GGCCATTAAC | TAATTAG | - |
| repo#MA0240.1 | -2 | 0.0062931 | 2.99552 | 0.0989725 | 7 | GGCCATTAAC | TAATTAA | - |
| CG32105#MA0178.1 | -2 | 0.006728 | 3.20253 | 0.102225 | 7 | GGCCATTAAC | TAATTAA | - |
| CG4328#MA0182.1 | -2 | 0.006728 | 3.20253 | 0.102225 | 7 | GGCCATTAAC | CAATAAA | - |
| cad#MA0216.1 | -2 | 0.00718606 | 3.42056 | 0.107365 | 7 | GGCCATTAAC | CAATAAA | - |
| Pdx1#MA0132.1 | -3 | 0.00761691 | 3.62565 | 0.110887 | 6 | GGCCATTAAC | AATTAG | - |
| CG9876#MA0184.1 | -2 | 0.00766916 | 3.65052 | 0.110887 | 7 | GGCCATTAAC | TAATTAG | - |
| HOXA5#MA0158.1 | -3 | 0.0109897 | 5.23111 | 0.156338 | 7 | GGCCATTAAC | AATTAGTG | - |
| Abd-B#MA0165.1 | -2 | 0.0111614 | 5.31285 | 0.156338 | 7 | GGCCATTAAC | TCATAAA | - |
| ARID3A#MA0151.1 | -4 | 0.0113447 | 5.40006 | 0.15646 | 6 | GGCCATTAAC | ATTAAA | + |
| inv#MA0229.1 | -2 | 0.0116203 | 5.53125 | 0.157039 | 8 | GGCCATTAAC | TAATTAGA | - |
| ro#MA0241.1 | -2 | 0.0118535 | 5.64227 | 0.157039 | 7 | GGCCATTAAC | TAATTAG | - |
| HAP5#MA0316.1 | 0 | 0.0119122 | 5.67022 | 0.157039 | 10 | GGCCATTAAC | ACCAATCAGATCCCA | - |
| Pph13#MA0200.1 | -2 | 0.0149831 | 7.13198 | 0.194661 | 7 | GGCCATTAAC | TAATTAG | - |
| YOX1#MA0433.1 | -1 | 0.0199051 | 9.47484 | 0.254913 | 8 | GGCCATTAAC | TTAATTAA | - |
| Lim1#MA0194.1 | -2 | 0.0209333 | 9.96426 | 0.264304 | 7 | GGCCATTAAC | TAATTAA | - |