MacroAPE 1083:3-MASTTCCCCTTW
From FANTOM5_SSTAR
Full Name: 3-MASTTCCCCTTW,CNhs11673,BestGuess:NFAT(RHD)/Jurkat-NFATC1-ChIP-Seq/Homer(0.739349270844042),T:141.0(4.14%),B:616.2(1.34%),P:1e-29
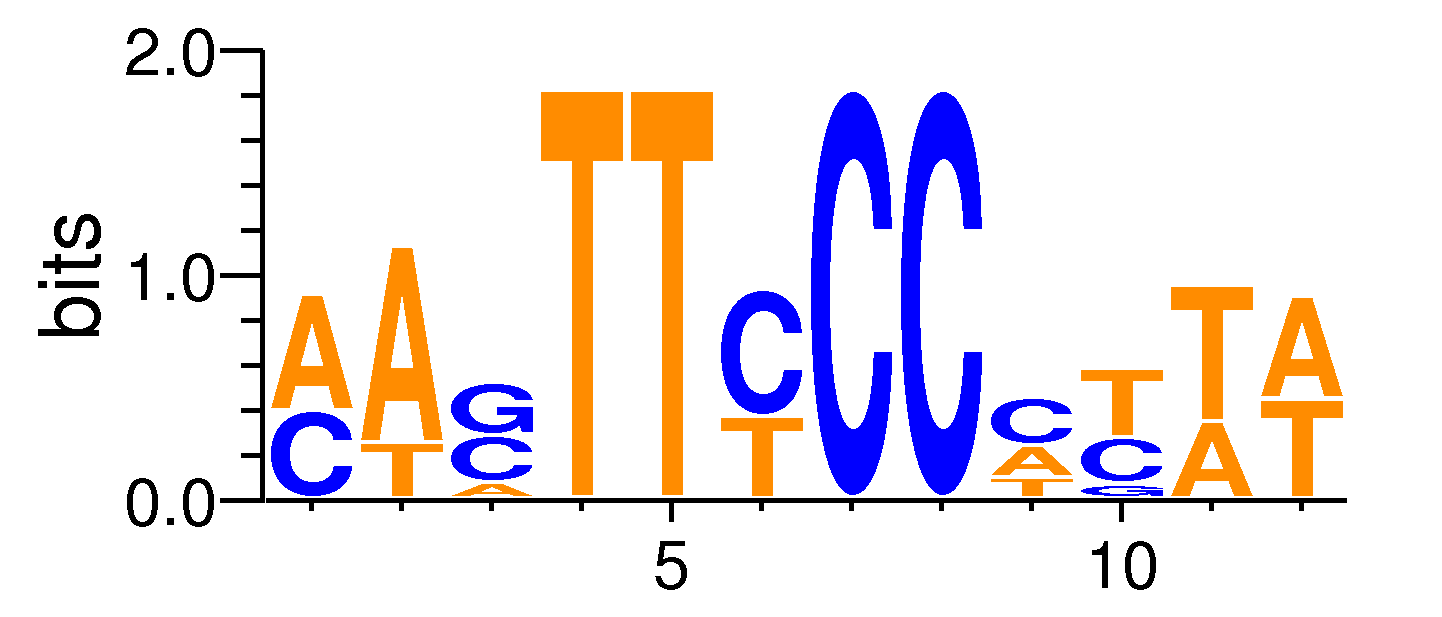
| ExpandMotif matrix |
|---|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| SPI1#MA0080.2 | -2 | 0.00100134 | 0.476638 | 0.811701 | 6 | AAGTTCCCCTTA | CTTCCG | - |
| SPI1#MA0080.2 | -1 | 0.00362538 | 1.72568 | 0.811701 | 7 | AAGTTCCCCTTA | ACTTCCT | - |
| MZF1_5-13#MA0057.1 | -3 | 0.00415742 | 1.97893 | 0.811701 | 9 | AAGTTCCCCTTA | TTCCCCCTAC | - |
| id1#MA0120.1 | -3 | 0.00440172 | 2.09522 | 0.811701 | 9 | AAGTTCCCCTTA | TTTTCCTTTTCG | + |
| STAT1#MA0137.1 | 0 | 0.00454865 | 2.16516 | 0.811701 | 12 | AAGTTCCCCTTA | CAGTTTCATTTTCC | - |
| squamosa#MA0082.1 | -2 | 0.00511706 | 2.43572 | 0.811701 | 10 | AAGTTCCCCTTA | ATTTCCATTTTTGG | - |
| Klf4#MA0039.1 | -1 | 0.00628837 | 2.99326 | 0.855001 | 10 | AAGTTCCCCTTA | CCTTCCTTTA | - |
| RELA#MA0107.1 | 2 | 0.0104658 | 4.98174 | 0.961545 | 8 | AAGTTCCCCTTA | GGAAATTCCC | - |
| NF-kappaB#MA0061.1 | 2 | 0.0116806 | 5.55998 | 0.961545 | 8 | AAGTTCCCCTTA | GGAAATTCCC | - |
| MZF1_1-4#MA0056.1 | -4 | 0.0133731 | 6.36561 | 0.961545 | 6 | AAGTTCCCCTTA | TCCCCA | - |
| SPIB#MA0081.1 | -3 | 0.0137473 | 6.5437 | 0.961545 | 7 | AAGTTCCCCTTA | TTCCTCT | - |
| USV1#MA0413.1 | 0 | 0.0154405 | 7.34966 | 0.961545 | 12 | AAGTTCCCCTTA | AAATTCCCCCTGAATTTGTG | + |
| IRF1#MA0050.1 | -1 | 0.015776 | 7.5094 | 0.961545 | 11 | AAGTTCCCCTTA | GGTTTCACTTTC | - |
| Spz1#MA0111.1 | 1 | 0.0162061 | 7.71409 | 0.961545 | 10 | AAGTTCCCCTTA | GCTGTTACCCT | - |
| Pax4#MA0068.1 | 3 | 0.0165354 | 7.87084 | 0.961545 | 12 | AAGTTCCCCTTA | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| znf143#MA0088.1 | 0 | 0.0171748 | 8.17521 | 0.961545 | 12 | AAGTTCCCCTTA | GATTTCCCATAATGCCTTGC | + |
| GIS1#MA0306.1 | -4 | 0.0203953 | 9.70817 | 0.992996 | 8 | AAGTTCCCCTTA | ACCCCTAAA | + |