MacroAPE 1083:8-CACACYCMCACA
From FANTOM5_SSTAR
Full Name: 8-CACACYCMCACA,CNhs11797,BestGuess:Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer(0.704632330650495),T:1055.0(25.62%),B:8135.8(18.35%),P:1e-30
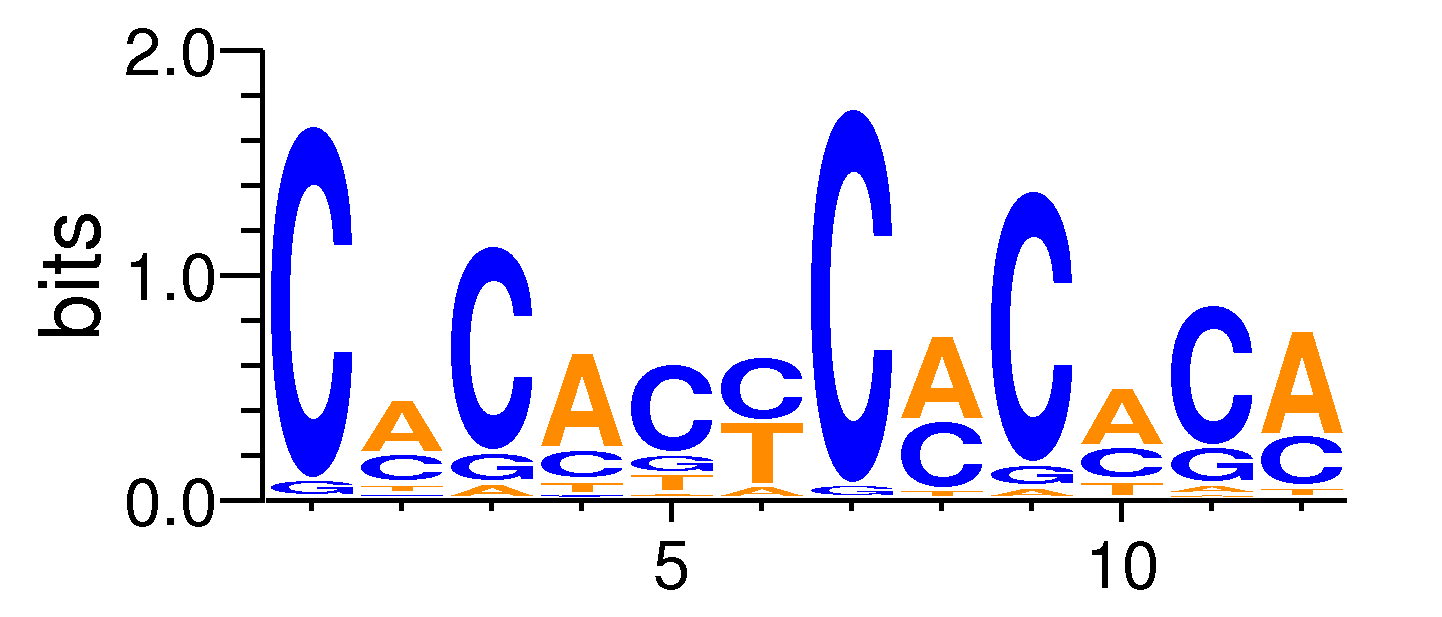
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| RREB1#MA0073.1 | 8 | 5.18389e-05 | 0.0246753 | 0.0490776 | 12 | CACACCCACACA | CCCCAAACCACCCCCCCCCC | + |
| Pax4#MA0068.1 | 16 | 0.000342584 | 0.16307 | 0.14993 | 12 | CACACCCACACA | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| RAP1#MA0359.1 | -2 | 0.000475095 | 0.226145 | 0.14993 | 10 | CACACCCACACA | CACCCATACA | + |
| AFT2#MA0270.1 | 0 | 0.00086826 | 0.413292 | 0.205503 | 8 | CACACCCACACA | CACACCCC | + |
| Egr1#MA0162.1 | -1 | 0.00135379 | 0.644406 | 0.256337 | 11 | CACACCCACACA | CCGCCCACGCA | - |
| CRZ1#MA0285.1 | 0 | 0.00173965 | 0.828076 | 0.274498 | 9 | CACACCCACACA | CTAAGCCAC | + |
| Klf4#MA0039.2 | 2 | 0.00228816 | 1.08916 | 0.309468 | 8 | CACACCCACACA | GCCCCACCCA | - |
| SP1#MA0079.2 | -1 | 0.00294705 | 1.4028 | 0.348759 | 10 | CACACCCACACA | ACCCCGCCCC | - |
| SP1#MA0079.2 | 1 | 0.00354978 | 1.68969 | 0.368239 | 9 | CACACCCACACA | CCCCGCCCCC | + |
| YGR067C#MA0425.1 | 2 | 0.00388957 | 1.85144 | 0.368239 | 12 | CACACCCACACA | ACCCCACTTTTTTG | + |
| Tal1::Gata1#MA0140.1 | 5 | 0.00506053 | 2.40881 | 0.435543 | 12 | CACACCCACACA | CTTATCTGTCCCCACCAG | - |
| btd#MA0443.1 | 0 | 0.00720341 | 3.42882 | 0.568309 | 10 | CACACCCACACA | TCCGCCCCCT | - |
| YPR022C#MA0436.1 | -5 | 0.0131082 | 6.2395 | 0.896277 | 7 | CACACCCACACA | CCCCACG | + |
| z#MA0255.1 | -3 | 0.0139217 | 6.62675 | 0.896277 | 9 | CACACCCACACA | AATCACTCAA | - |
| hkb#MA0450.1 | 1 | 0.0142006 | 6.75947 | 0.896277 | 8 | CACACCCACACA | TCACGCCCC | - |
| ESR1#MA0112.2 | 6 | 0.0181167 | 8.62355 | 0.920075 | 12 | CACACCCACACA | CCAGGTCACCGTGACCCC | + |
| ESR2#MA0258.1 | 5 | 0.0190055 | 9.04661 | 0.920075 | 12 | CACACCCACACA | CAGGTCACCGTGACCTTG | - |
| INSM1#MA0155.1 | 0 | 0.019278 | 9.17632 | 0.920075 | 12 | CACACCCACACA | CGCCCCCTGACA | - |
| MIG1#MA0337.1 | -4 | 0.0194767 | 9.27091 | 0.920075 | 7 | CACACCCACACA | CCCCCGC | + |
| PPARG#MA0066.1 | 6 | 0.0204087 | 9.71453 | 0.920075 | 12 | CACACCCACACA | GTAGGTCACGGTGACCTACT | + |
| ESR1#MA0112.2 | 9 | 0.0204087 | 9.71453 | 0.920075 | 11 | CACACCCACACA | GGCCCAGGTCACCCTGACCT | + |