MacroAPE 1083:CNhs11185 bl f2
From FANTOM5_SSTAR
Full Name: CNhs11185_bl_f2
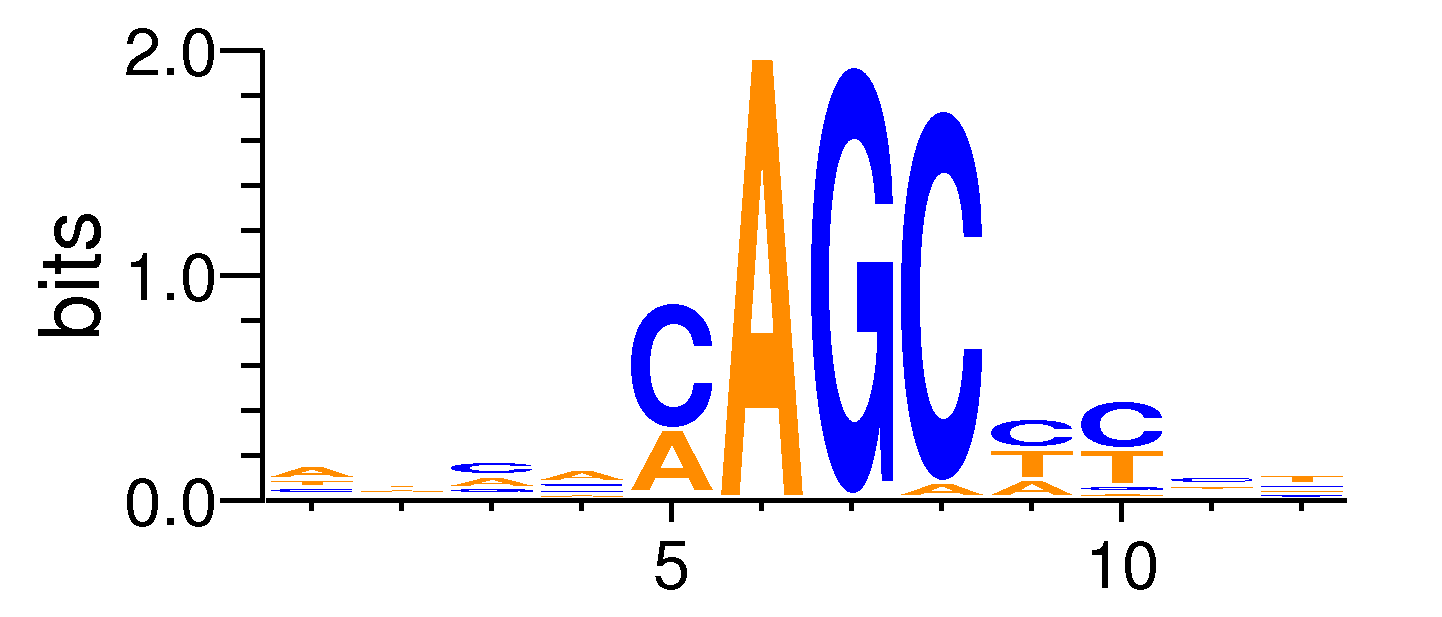
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| CRZ1#MA0285.1 | -2 | 0.000305441 | 0.14539 | 0.29078 | 9 | AACACAGCCCCT | CTAAGCCAC | + |
| ACE2#MA0267.1 | -2 | 0.00327887 | 1.56074 | 0.797602 | 7 | AACACAGCCCCT | ACCAGCA | + |
| SWI5#MA0402.1 | -1 | 0.00336202 | 1.60032 | 0.797602 | 8 | AACACAGCCCCT | AACCAGCA | - |
| SP1#MA0079.1 | -1 | 0.00359362 | 1.71056 | 0.797602 | 10 | AACACAGCCCCT | ACCCCGCCCC | - |
| Dof2#MA0020.1 | -3 | 0.00601921 | 2.86514 | 0.797602 | 6 | AACACAGCCCCT | AAAGCA | + |
| Pax4#MA0068.1 | 14 | 0.00606848 | 2.8886 | 0.797602 | 12 | AACACAGCCCCT | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| Pax5#MA0014.1 | 9 | 0.00662935 | 3.15557 | 0.797602 | 11 | AACACAGCCCCT | CCGCTACGCTTCAGTGCTCT | - |
| TOD6#MA0350.1 | 1 | 0.00670254 | 3.19041 | 0.797602 | 12 | AACACAGCCCCT | AGGCACAGCTCATCGCGTTTT | + |
| Mafb#MA0117.1 | 0 | 0.0092951 | 4.42447 | 0.983215 | 8 | AACACAGCCCCT | GCGTCAGC | - |
| NHLH1#MA0048.1 | -1 | 0.0151245 | 7.19924 | 1 | 11 | AACACAGCCCCT | GCGCAGCTGCGT | + |
| MNB1A#MA0053.1 | -3 | 0.0167376 | 7.96708 | 1 | 5 | AACACAGCCCCT | AAAGC | + |
| DOT6#MA0351.1 | 1 | 0.0176647 | 8.40841 | 1 | 12 | AACACAGCCCCT | TTCTGCACCTCATCGCATCCT | + |
| GAL4#MA0299.1 | 3 | 0.0196715 | 9.36364 | 1 | 12 | AACACAGCCCCT | CGGGGAACAGTACTC | + |
| PBF#MA0064.1 | -3 | 0.0201853 | 9.6082 | 1 | 5 | AACACAGCCCCT | AAAGC | + |