MacroAPE 1083:CNhs12105 bl f2
From FANTOM5_SSTAR
Full Name: CNhs12105_bl_f2
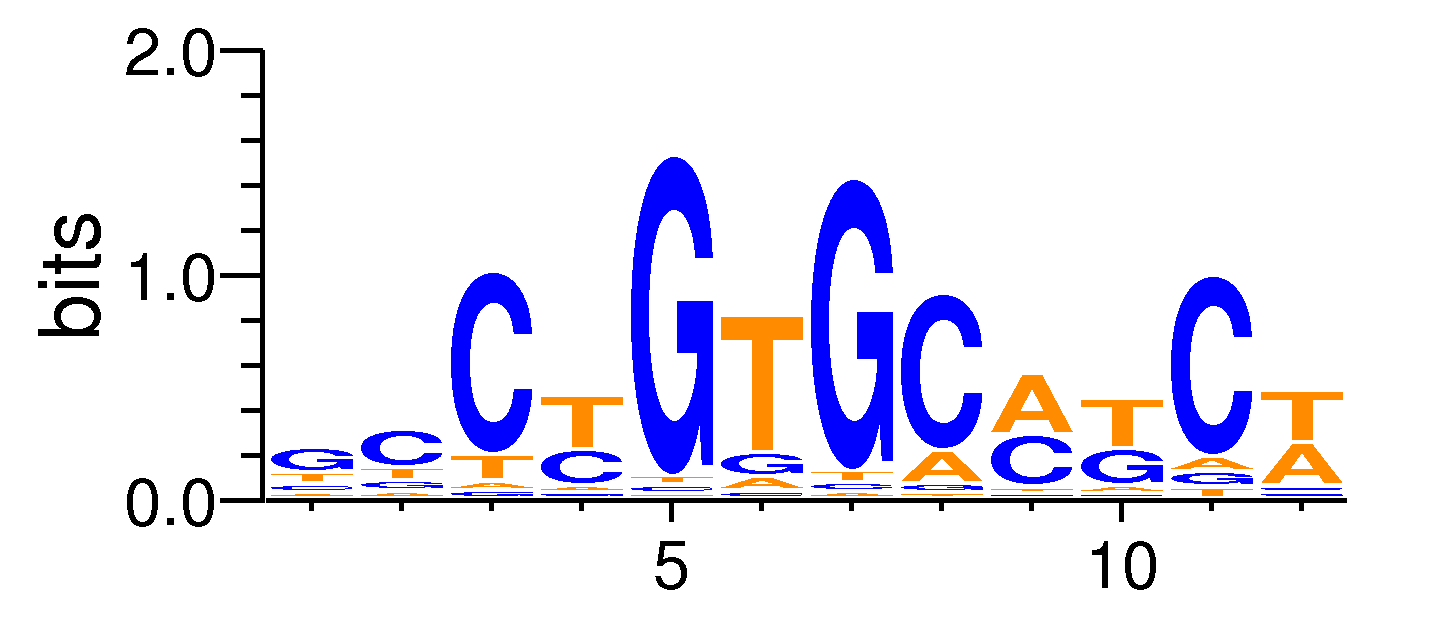
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| PHD1#MA0355.1 | -2 | 0.00513112 | 2.44241 | 0.75512 | 10 | GCCTGTGCATCT | TGCTGCAGGT | - |
| HIF1A::ARNT#MA0259.1 | 0 | 0.0071099 | 3.38431 | 0.75512 | 8 | GCCTGTGCATCT | GGACGTGC | + |
| TP53#MA0106.1 | 8 | 0.00747541 | 3.5583 | 0.75512 | 12 | GCCTGTGCATCT | CCGGACATGCCCGGGCATGT | + |
| PHO4#MA0357.1 | 0 | 0.00752759 | 3.58313 | 0.75512 | 8 | GCCTGTGCATCT | GCACGTGC | + |
| Zfx#MA0146.1 | 3 | 0.0082011 | 3.90372 | 0.75512 | 11 | GCCTGTGCATCT | GGGGCCGAGGCCTG | + |
| h#MA0449.1 | 1 | 0.00836445 | 3.98148 | 0.75512 | 9 | GCCTGTGCATCT | GGCACGTGCC | + |
| SOK2#MA0385.1 | -1 | 0.00873885 | 4.15969 | 0.75512 | 11 | GCCTGTGCATCT | TGCCTGCAGGT | - |
| RTG3#MA0376.1 | 6 | 0.0112288 | 5.34491 | 0.865634 | 12 | GCCTGTGCATCT | ATATTAGCACGTGCTTAGTC | + |
| MET28#MA0332.1 | -2 | 0.0118392 | 5.63547 | 0.865634 | 6 | GCCTGTGCATCT | CTGTGG | + |
| INO2#MA0321.1 | 0 | 0.0130555 | 6.2144 | 0.886377 | 9 | GCCTGTGCATCT | GCATGTGAA | + |
| RUNX1#MA0002.2 | 0 | 0.0157687 | 7.5059 | 0.919175 | 11 | GCCTGTGCATCT | GTCTGTGGTTT | + |
| INO4#MA0322.1 | 0 | 0.0160409 | 7.63548 | 0.919175 | 9 | GCCTGTGCATCT | GCATGTGAA | + |
| Ar#MA0007.1 | 9 | 0.0186248 | 8.86541 | 0.919175 | 12 | GCCTGTGCATCT | ATAAGAACACCCTGTACCCGCC | + |
| CBF1#MA0281.1 | 0 | 0.0190643 | 9.07461 | 0.919175 | 8 | GCCTGTGCATCT | TCACGTGC | - |
| HNF4A#MA0114.1 | 2 | 0.0193408 | 9.20621 | 0.919175 | 11 | GCCTGTGCATCT | TGACCTTTGGCCT | - |
| abi4#MA0123.1 | -2 | 0.0206804 | 9.84388 | 0.936041 | 10 | GCCTGTGCATCT | CGGTGCCCCC | + |