MacroAPE 1083:CNhs12920 bl f2
From FANTOM5_SSTAR
Full Name: CNhs12920_bl_f2
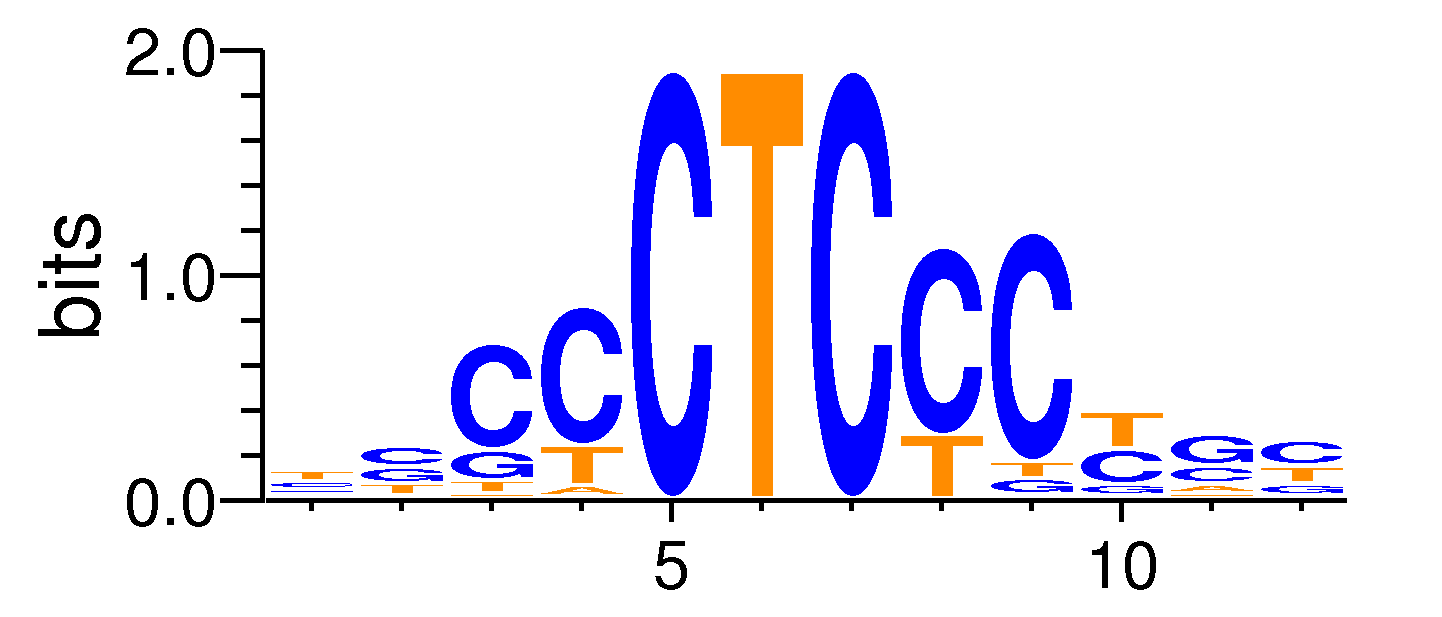
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| SP1#MA0079.2 | -1 | 9.52054e-05 | 0.0453178 | 0.0905169 | 10 | TCCCCTCCCTGC | CCCCGCCCCC | + |
| YGR067C#MA0425.1 | 0 | 0.00040282 | 0.191742 | 0.191491 | 12 | TCCCCTCCCTGC | ACCCCACTTTTTTG | + |
| Trl#MA0205.1 | 1 | 0.00158806 | 0.755919 | 0.390529 | 9 | TCCCCTCCCTGC | TTGCTCTCTC | + |
| Pax4#MA0068.1 | 16 | 0.00195562 | 0.930875 | 0.390529 | 12 | TCCCCTCCCTGC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| YML081W#MA0431.1 | 0 | 0.00205379 | 0.977603 | 0.390529 | 9 | TCCCCTCCCTGC | ACCCCGCAC | + |
| RXR::RAR_DR5#MA0159.1 | 0 | 0.00276723 | 1.3172 | 0.438493 | 12 | TCCCCTCCCTGC | TGACCTCTCCGTGACCT | - |
| btd#MA0443.1 | -2 | 0.00529478 | 2.52032 | 0.65776 | 10 | TCCCCTCCCTGC | TCCGCCCCCT | - |
| Klf4#MA0039.2 | -2 | 0.00589751 | 2.80721 | 0.65776 | 10 | TCCCCTCCCTGC | CCTTCCTTTA | - |
| Klf4#MA0039.2 | 0 | 0.0069183 | 3.29311 | 0.65776 | 10 | TCCCCTCCCTGC | GCCCCACCCA | - |
| MZF1_5-13#MA0057.1 | 2 | 0.0069183 | 3.29311 | 0.65776 | 8 | TCCCCTCCCTGC | TTCCCCCTAC | - |
| opa#MA0456.1 | -2 | 0.00988634 | 4.7059 | 0.663301 | 10 | TCCCCTCCCTGC | GACCCCCCGCTG | + |
| SP1#MA0079.2 | 0 | 0.00994628 | 4.73443 | 0.663301 | 10 | TCCCCTCCCTGC | ACCCCGCCCC | - |
| GABPA#MA0062.2 | 0 | 0.0100117 | 4.76556 | 0.663301 | 11 | TCCCCTCCCTGC | GCCACTTCCGG | - |
| RGM1#MA0366.1 | -1 | 0.0103393 | 4.92151 | 0.663301 | 5 | TCCCCTCCCTGC | CCCCT | - |
| GABPA#MA0062.2 | -2 | 0.0104649 | 4.98128 | 0.663301 | 10 | TCCCCTCCCTGC | CTCTTCCGGT | - |
| IXR1#MA0323.1 | 1 | 0.0121009 | 5.76001 | 0.719059 | 12 | TCCCCTCCCTGC | CCCCGCTTCCGGCTT | - |
| Ar#MA0007.1 | 9 | 0.0149144 | 7.09924 | 0.834112 | 12 | TCCCCTCCCTGC | ATAAGAACACCCTGTACCCGCC | + |
| MZF1_1-4#MA0056.1 | -5 | 0.0163891 | 7.80121 | 0.865667 | 6 | TCCCCTCCCTGC | TCCCCA | - |
| MSN4#MA0342.1 | -1 | 0.0192318 | 9.15434 | 0.948788 | 5 | TCCCCTCCCTGC | CCCCT | - |
| UGA3#MA0410.1 | -1 | 0.0199587 | 9.50032 | 0.948788 | 8 | TCCCCTCCCTGC | TCCCGCCG | - |