MacroAPE 1083:INSM1 f1
From FANTOM5_SSTAR
Full Name: INSM1_f1
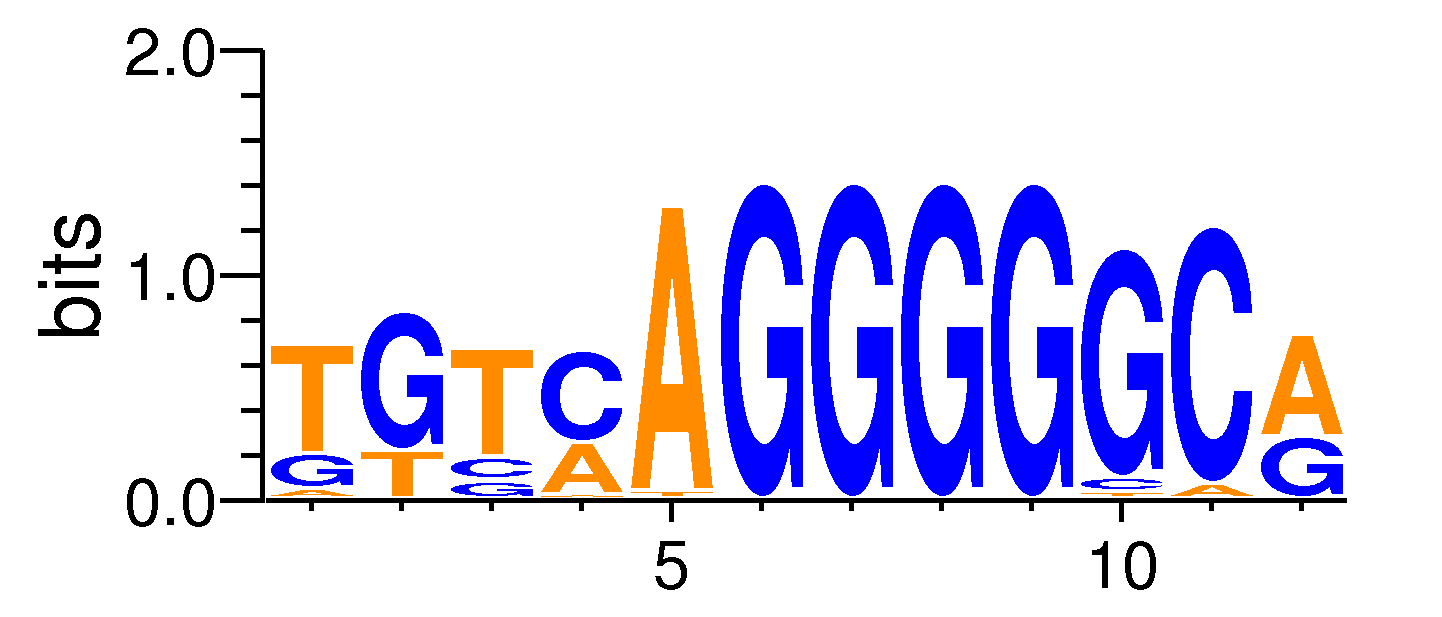
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| INSM1#MA0155.1 | 0 | 1.4344e-10 | 6.82773e-08 | 1.35078e-07 | 12 | TGTCAGGGGGCA | TGTCAGGGGGCG | + |
| REI1#MA0364.1 | -2 | 0.000104977 | 0.0499692 | 0.0377484 | 7 | TGTCAGGGGGCA | TCAGGGG | - |
| USV1#MA0413.1 | 5 | 0.000120255 | 0.0572414 | 0.0377484 | 12 | TGTCAGGGGGCA | CACAAATTCAGGGGGAATTT | - |
| MSN4#MA0342.1 | -4 | 0.000215039 | 0.102359 | 0.0487956 | 5 | TGTCAGGGGGCA | AGGGG | + |
| RGM1#MA0366.1 | -4 | 0.00025908 | 0.123322 | 0.0487956 | 5 | TGTCAGGGGGCA | AGGGG | + |
| MSN2#MA0341.1 | -4 | 0.000523039 | 0.248967 | 0.0820918 | 5 | TGTCAGGGGGCA | AGGGG | + |
| btd#MA0443.1 | -4 | 0.000820773 | 0.390688 | 0.110418 | 8 | TGTCAGGGGGCA | AGGGGGCGGA | + |
| GIS1#MA0306.1 | -1 | 0.00101981 | 0.485427 | 0.120045 | 9 | TGTCAGGGGGCA | TTTAGGGGT | - |
| RPH1#MA0372.1 | -2 | 0.00208064 | 0.990386 | 0.217707 | 8 | TGTCAGGGGGCA | TTAGGGGT | - |
| ceh-22#MA0264.1 | 0 | 0.00233214 | 1.1101 | 0.219619 | 11 | TGTCAGGGGGCA | TTTCAAGTGGT | - |
| abi4#MA0123.1 | -5 | 0.00322153 | 1.53345 | 0.267814 | 7 | TGTCAGGGGGCA | GGGGGCACCG | - |
| PPARG::RXRA#MA0065.2 | 3 | 0.00352485 | 1.67783 | 0.267814 | 12 | TGTCAGGGGGCA | GTAGGGCAAAGGTCA | + |
| MZF1_5-13#MA0057.1 | -2 | 0.00369709 | 1.75982 | 0.267814 | 10 | TGTCAGGGGGCA | GGAGGGGGAA | + |
| Macho-1#MA0118.1 | -3 | 0.00437819 | 2.08402 | 0.294498 | 9 | TGTCAGGGGGCA | TGGGGGGTC | + |
| RPN4#MA0373.1 | -5 | 0.00496756 | 2.36456 | 0.306257 | 7 | TGTCAGGGGGCA | GGTGGCG | + |
| YER130C#MA0423.1 | -1 | 0.00551844 | 2.62678 | 0.306257 | 9 | TGTCAGGGGGCA | AATAGGGGT | - |
| RAP1#MA0359.1 | 0 | 0.00552864 | 2.63163 | 0.306257 | 10 | TGTCAGGGGGCA | TGTATGGGTG | - |
| NRG1#MA0347.1 | 3 | 0.00605719 | 2.88322 | 0.309354 | 12 | TGTCAGGGGGCA | CTAGATCAGGGTCCATCGCA | + |
| CTCF#MA0139.1 | 4 | 0.00624156 | 2.97098 | 0.309354 | 12 | TGTCAGGGGGCA | TGGCCACCAGGGGGCGCTA | + |
| SP1#MA0079.2 | -5 | 0.00781094 | 3.71801 | 0.367782 | 7 | TGTCAGGGGGCA | GGGGGCGGGG | - |
| HNF4A#MA0114.1 | 1 | 0.00919448 | 4.37657 | 0.412311 | 12 | TGTCAGGGGGCA | AGGCCAAAGGTCA | + |
| YML081W#MA0431.1 | -1 | 0.0122442 | 5.82822 | 0.524111 | 9 | TGTCAGGGGGCA | GTGCGGGGT | - |
| PPARG::RXRA#MA0065.2 | -2 | 0.0132785 | 6.32059 | 0.543675 | 10 | TGTCAGGGGGCA | CTAGGGGTCAAAGGTCATCG | + |
| opa#MA0456.1 | 1 | 0.0171751 | 8.17535 | 0.667386 | 11 | TGTCAGGGGGCA | CAGCGGGGGGTC | - |
| YJL103C#MA0427.1 | 0 | 0.0185831 | 8.84557 | 0.667386 | 11 | TGTCAGGGGGCA | TCTCCGGAGCT | - |
| h#MA0449.1 | -1 | 0.0191348 | 9.10818 | 0.667386 | 10 | TGTCAGGGGGCA | GGCACGTGCC | + |