MacroAPE 1083:PAX4
From FANTOM5_SSTAR
Full Name: PAX4
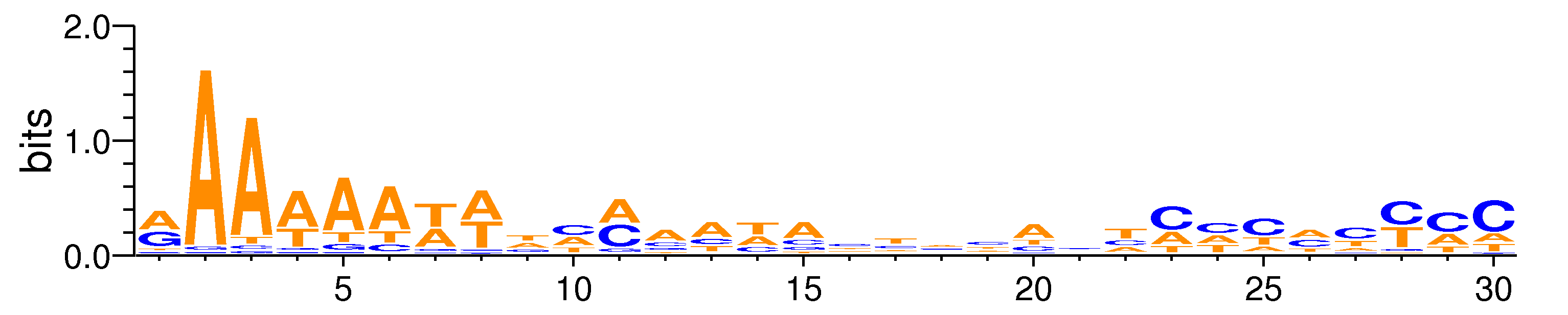
| Motif matrix | |
|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Pax4#MA0068.1 | 0 | 0 | 0 | 0 | 30 | AAAAAATATCAAATACTACACTCCCACCCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| SUM1#MA0398.1 | -1 | 2.17753e-05 | 0.010365 | 0.0092566 | 9 | AAAAAATATCAAATACTACACTCCCACCCC | AAAAATTTT | - |
| HMG-I/Y#MA0045.1 | 0 | 2.95518e-05 | 0.0140667 | 0.0092566 | 16 | AAAAAATATCAAATACTACACTCCCACCCC | CAACAAATGGAAAAAC | + |
| MGA1#MA0336.1 | -1 | 0.000100708 | 0.0479369 | 0.0236587 | 21 | AAAAAATATCAAATACTACACTCCCACCCC | AGGACTATAGAACACTCTAAA | + |
| SFP1#MA0378.1 | 5 | 0.000363626 | 0.173086 | 0.0599899 | 16 | AAAAAATATCAAATACTACACTCCCACCCC | CCGTAGAAAATTTTTTTCAAT | - |
| NHP6A#MA0345.1 | 0 | 0.000386696 | 0.184067 | 0.0599899 | 21 | AAAAAATATCAAATACTACACTCCCACCCC | TCATTTTTATATATAGGTCAT | - |
| NHP6B#MA0346.1 | 0 | 0.000446877 | 0.212713 | 0.0599899 | 20 | AAAAAATATCAAATACTACACTCCCACCCC | TCATATTATATATAAATAAA | - |
| STB3#MA0390.1 | 7 | 0.000705994 | 0.336053 | 0.0829276 | 14 | AAAAAATATCAAATACTACACTCCCACCCC | CCAGAGTGAAAAATTTTGGAC | - |
| EDS1#MA0294.1 | 2 | 0.00122849 | 0.584762 | 0.104392 | 7 | AAAAAATATCAAATACTACACTCCCACCCC | CGGAAAAAT | + |
| ARO80#MA0273.1 | 0 | 0.0013331 | 0.634553 | 0.104392 | 21 | AAAAAATATCAAATACTACACTCCCACCCC | ACATACTTGCCGAGAATTATC | - |
| SFL1#MA0377.1 | 1 | 0.00208182 | 0.990946 | 0.139734 | 20 | AAAAAATATCAAATACTACACTCCCACCCC | TAGAGAATAGAAGAAATAAAA | + |
| RGT1#MA0367.1 | 3 | 0.00297253 | 1.41493 | 0.186219 | 8 | AAAAAATATCAAATACTACACTCCCACCCC | CCGGAAAAACT | - |
| br_Z1#MA0010.1 | -2 | 0.00401736 | 1.91226 | 0.235944 | 14 | AAAAAATATCAAATACTACACTCCCACCCC | GTAATAAACAAATC | + |
| hb#MA0049.1 | 4 | 0.00465809 | 2.21725 | 0.257482 | 6 | AAAAAATATCAAATACTACACTCCCACCCC | GCATAAAAAA | + |
| squamosa#MA0082.1 | 1 | 0.00695244 | 3.30936 | 0.343852 | 13 | AAAAAATATCAAATACTACACTCCCACCCC | CCAAAAATGGAAAT | + |
| TBP#MA0386.1 | 0 | 0.00770996 | 3.66994 | 0.362252 | 21 | AAAAAATATCAAATACTACACTCCCACCCC | GACTAGATATATATATTCGAT | + |
| br_Z4#MA0013.1 | 4 | 0.00893364 | 4.25241 | 0.399758 | 7 | AAAAAATATCAAATACTACACTCCCACCCC | TAGTAAACAAA | + |
| HAT5#MA0008.1 | -1 | 0.0113421 | 5.39882 | 0.467031 | 8 | AAAAAATATCAAATACTACACTCCCACCCC | AATAATTG | - |
| SMP1#MA0383.1 | -6 | 0.011431 | 5.44117 | 0.467031 | 21 | AAAAAATATCAAATACTACACTCCCACCCC | TTACCTATAATTAAATTAGCA | + |
| br_Z3#MA0012.1 | -1 | 0.0150861 | 7.18099 | 0.521685 | 11 | AAAAAATATCAAATACTACACTCCCACCCC | TAAACTAAAAG | + |
| FKH1#MA0296.1 | 1 | 0.0157283 | 7.48667 | 0.521685 | 19 | AAAAAATATCAAATACTACACTCCCACCCC | TGAATTGTAAACAAAGAGGA | + |
| TBP#MA0386.1 | 0 | 0.0166549 | 7.92772 | 0.521685 | 15 | AAAAAATATCAAATACTACACTCCCACCCC | GTATAAAAGGCGGGG | + |
| TBP#MA0386.1 | 0 | 0.0166549 | 7.92772 | 0.521685 | 15 | AAAAAATATCAAATACTACACTCCCACCCC | GTATAAAAGGCGGGG | + |
| FOXI1#MA0042.1 | 0 | 0.0176939 | 8.4223 | 0.536353 | 12 | AAAAAATATCAAATACTACACTCCCACCCC | AAACAAACATCC | - |