MacroAPE 1083:RARA f1
From FANTOM5_SSTAR
Full Name: RARA_f1
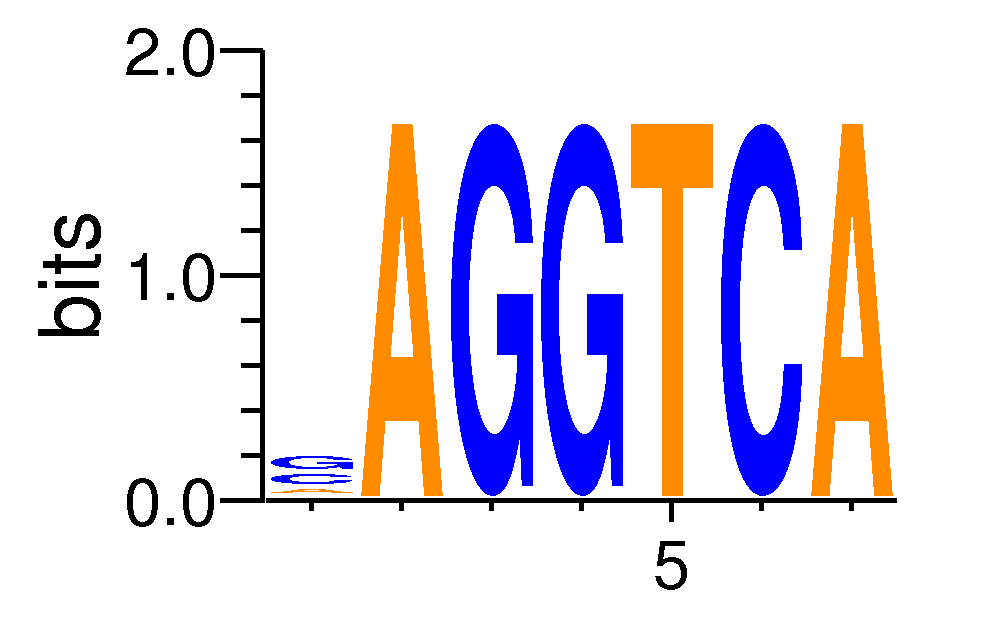
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| RXR::RAR_DR5#MA0159.1 | 10 | 3.75373e-05 | 0.0178677 | 0.0258198 | 7 | GAGGTCA | AGGTCATGGAGAGGTCA | + |
| RORA_1#MA0071.1 | 3 | 7.51396e-05 | 0.0357665 | 0.0258198 | 7 | GAGGTCA | ATCAAGGTCA | + |
| NR4A2#MA0160.1 | 0 | 8.29026e-05 | 0.0394616 | 0.0258198 | 7 | GAGGTCA | AAGGTCAC | + |
| ESR1#MA0112.2 | 4 | 0.000207565 | 0.0988011 | 0.0435101 | 7 | GAGGTCA | GGCCCAGGTCACCCTGACCT | + |
| ESR2#MA0258.1 | 0 | 0.000251598 | 0.119761 | 0.0435101 | 7 | GAGGTCA | CAGGTCACCGTGACCTTG | - |
| ESR1#MA0112.2 | 1 | 0.000279405 | 0.132997 | 0.0435101 | 7 | GAGGTCA | CCAGGTCACCGTGACCCC | + |
| NR1H2::RXRA#MA0115.1 | 1 | 0.000527749 | 0.251208 | 0.0704426 | 7 | GAGGTCA | AAAGGTCAAAGGTCAAC | + |
| Esrrb#MA0141.1 | 5 | 0.000648864 | 0.308859 | 0.0757827 | 7 | GAGGTCA | AGCTCAAGGTCA | + |
| PPARG::RXRA#MA0065.2 | 8 | 0.000758351 | 0.360975 | 0.078729 | 7 | GAGGTCA | GTAGGGCAAAGGTCA | + |
| PPARG::RXRA#MA0065.2 | 3 | 0.00162121 | 0.771698 | 0.126231 | 7 | GAGGTCA | CTAGGGGTCAAAGGTCATCG | + |
| NR2F1#MA0017.1 | 7 | 0.00257667 | 1.22649 | 0.185192 | 7 | GAGGTCA | AGGTTCAAAGGTCA | - |
| CREB1#MA0018.2 | 1 | 0.00294631 | 1.40244 | 0.196633 | 7 | GAGGTCA | TGACGTCA | + |
| RORA_2#MA0072.1 | 6 | 0.00322306 | 1.53418 | 0.200763 | 7 | GAGGTCA | TATAAGTAGGTCAA | + |
| HNF4A#MA0114.1 | 6 | 0.0046296 | 2.20369 | 0.240313 | 7 | GAGGTCA | AGGCCAAAGGTCA | + |
| bZIP910#MA0096.1 | -1 | 0.00515903 | 2.4557 | 0.243571 | 6 | GAGGTCA | ACGTCAT | - |
| RXRA::VDR#MA0074.1 | -1 | 0.00521373 | 2.48174 | 0.243571 | 6 | GAGGTCA | GGGTCAACGAGTTCA | + |
| ZAP1#MA0440.1 | 6 | 0.0061139 | 2.91021 | 0.269466 | 7 | GAGGTCA | ACCTTAAAGGTCATG | + |
| PPARG#MA0066.1 | 1 | 0.00634483 | 3.02014 | 0.269466 | 7 | GAGGTCA | GTAGGTCACGGTGACCTACT | + |
| TGA1A#MA0129.1 | 0 | 0.00718569 | 3.42039 | 0.282382 | 7 | GAGGTCA | TACGTCA | + |
| usp#MA0016.1 | 0 | 0.0072534 | 3.45262 | 0.282382 | 7 | GAGGTCA | GGGGTCACGG | + |
| CST6#MA0286.1 | 1 | 0.00803937 | 3.82674 | 0.300461 | 7 | GAGGTCA | TTACGTCAT | - |
| CREB1#MA0018.2 | 0 | 0.00958928 | 4.5645 | 0.335082 | 7 | GAGGTCA | GACGTCACCAGG | - |
| bZIP911#MA0097.1 | 3 | 0.0161822 | 7.70273 | 0.539991 | 7 | GAGGTCA | GGCCACGTCATC | - |
| hth#MA0227.1 | -1 | 0.0187441 | 8.92218 | 0.603911 | 6 | GAGGTCA | CTGTCA | - |
| tll#MA0459.1 | 1 | 0.021 | 9.99598 | 0.654039 | 7 | GAGGTCA | AAAAGTCAAA | + |