MacroAPE 1083:RREB1 si
From FANTOM5_SSTAR
Full Name: RREB1_si
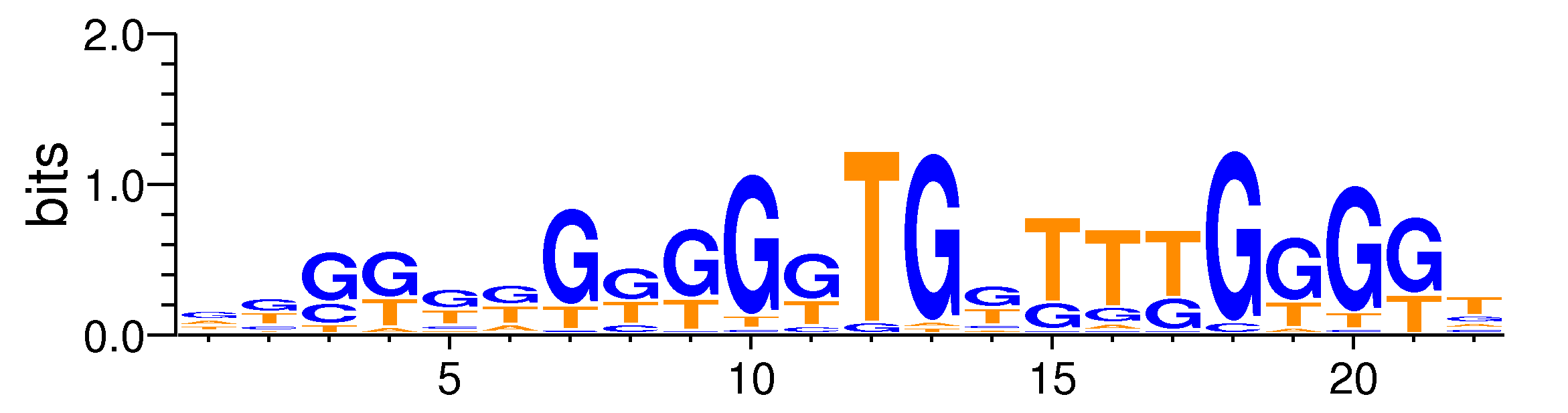
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| RREB1#MA0073.1 | -1 | 2.49717e-34 | 1.18865e-31 | 2.37731e-31 | 20 | GGGGGGGGGGGTGGTTTGGGGT | TGGGGGGGGGTGGTTTGGGG | - |
| Pax4#MA0068.1 | 1 | 5.99012e-07 | 0.00028513 | 0.00028513 | 22 | GGGGGGGGGGGTGGTTTGGGGT | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| ADR1#MA0268.1 | -15 | 0.00157934 | 0.751767 | 0.501178 | 7 | GGGGGGGGGGGTGGTTTGGGGT | GTGGGGT | - |
| YGR067C#MA0425.1 | -8 | 0.00477413 | 2.27249 | 0.859624 | 14 | GGGGGGGGGGGTGGTTTGGGGT | TGAAAAAGTGGGGT | - |
| HAP4#MA0315.1 | -4 | 0.00566648 | 2.69724 | 0.859624 | 15 | GGGGGGGGGGGTGGTTTGGGGT | TCGCCTCTGATTGGT | - |
| RUNX1#MA0002.2 | -6 | 0.00648601 | 3.08734 | 0.859624 | 11 | GGGGGGGGGGGTGGTTTGGGGT | GTCTGTGGTTT | + |
| Macho-1#MA0118.1 | -4 | 0.00754096 | 3.5895 | 0.859624 | 9 | GGGGGGGGGGGTGGTTTGGGGT | TGGGGGGTC | + |
| RPH1#MA0372.1 | -14 | 0.00781245 | 3.71872 | 0.859624 | 8 | GGGGGGGGGGGTGGTTTGGGGT | TTAGGGGT | - |
| SP1#MA0079.1 | -6 | 0.010211 | 4.86042 | 0.859624 | 10 | GGGGGGGGGGGTGGTTTGGGGT | GGGGCGGGGT | + |
| ESR1#MA0112.2 | -2 | 0.010607 | 5.04892 | 0.859624 | 20 | GGGGGGGGGGGTGGTTTGGGGT | AGGTCAGGGTGACCTGGGCC | - |
| BRCA1#MA0133.1 | -12 | 0.0109376 | 5.20629 | 0.859624 | 7 | GGGGGGGGGGGTGGTTTGGGGT | GTGTTGT | - |
| Gfi#MA0038.1 | -8 | 0.0112731 | 5.366 | 0.859624 | 10 | GGGGGGGGGGGTGGTTTGGGGT | CAGTGATTTG | - |
| SWI5#MA0402.1 | -8 | 0.0125731 | 5.98477 | 0.859624 | 8 | GGGGGGGGGGGTGGTTTGGGGT | TGCTGGTT | + |
| AFT1#MA0269.1 | -2 | 0.0130466 | 6.21018 | 0.859624 | 20 | GGGGGGGGGGGTGGTTTGGGGT | CCAATCGGGTGCAATATGTAA | - |
| NDT80#MA0343.1 | 0 | 0.0135445 | 6.44718 | 0.859624 | 21 | GGGGGGGGGGGTGGTTTGGGGT | TTGCGTTTTTGTGGCCGGAAA | - |
| AFT2#MA0270.1 | -7 | 0.016696 | 7.94729 | 0.993412 | 8 | GGGGGGGGGGGTGGTTTGGGGT | GGGGTGTG | - |
| INSM1#MA0155.1 | -11 | 0.0180609 | 8.59697 | 1 | 11 | GGGGGGGGGGGTGGTTTGGGGT | TGTCAGGGGGCG | + |
| Gamyb#MA0034.1 | -9 | 0.0200171 | 9.52816 | 1 | 10 | GGGGGGGGGGGTGGTTTGGGGT | GGCGGTTGTC | - |